| ||||
| SKBR3 PacBio Sequencing and Assembly Cold Spring Harbor Laboratory and Ontario Institute for Cancer Research
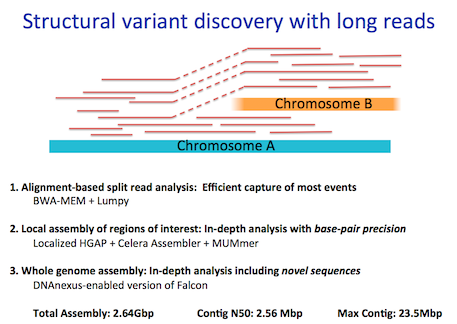
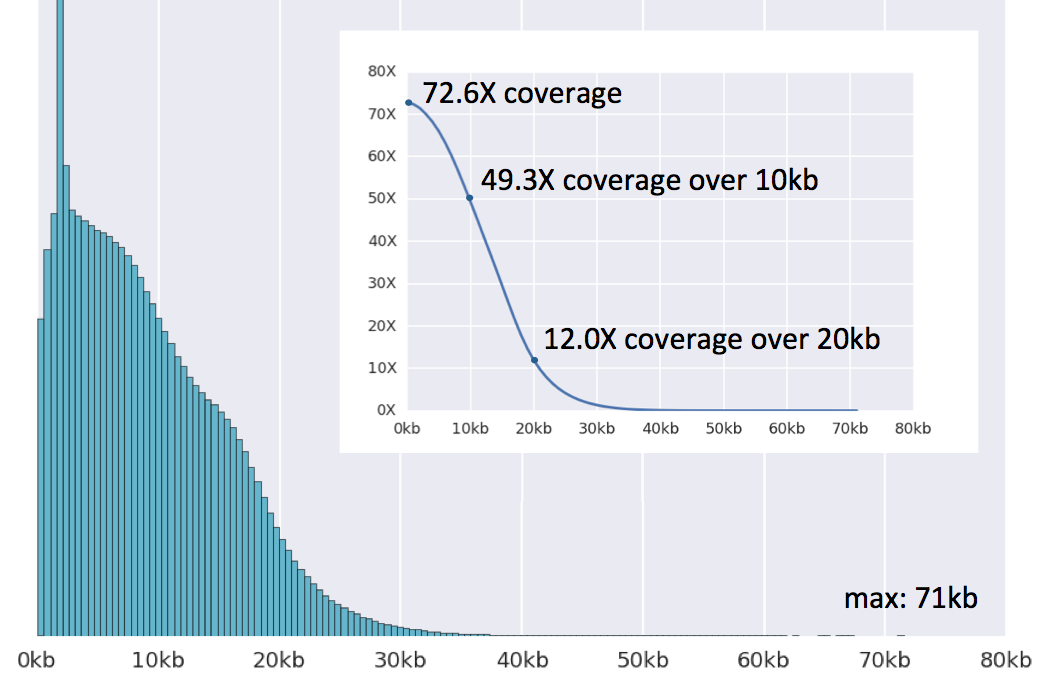
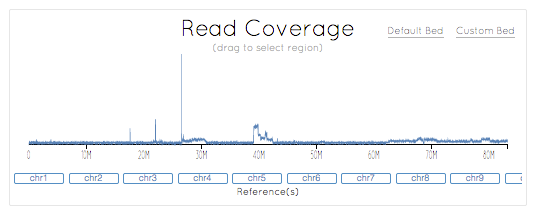
To address these challenges, we have sequenced SK-BR-3 using PacBio long read technology. Using the new P6-C4 chemistry, we generated more than 70x coverage of the genome with average read lengths of 9-13kb (max: 71kb). PacBio read coverage is highly correlated with the copy number assignments made using short read sequencing technologies, although the long reads provide more consistent coverage across repetitive elements. Furthermore, using the structural variation analysis program LUMPY and our new hybrid mapping and de novo assembly algorithm for analyzing split-read alignments, we have developed a detailed map of structural variations in this cell line. We have tentatively identified more than 900 intra-chromosomal and 300 inter-chromosomal variations, including many of the previously known gene fusions in SK-BR-3. Taking advantage of the newly identified breakpoints, we have developed an algorithm to reconstruct the mutational history of this cancer genome. From this we have characterized the amplifications of the HER2 region, discovering a complex series of nested duplications and translocations between chr17 and chr8, two of the most frequent translocation partners in primary breast cancers. To our knowledge, this establishes the most complete cancer reference genome to date.
Data Usage Agreement Users of these for genome wide analysis prior to our publication must agree to co-authorship as specified by the Toronto agreement.
By clicking these links, you agree to the Toronto agreement: |