Preprints
|
| P6. |
LRSim: a Linked Reads Simulator generating insights for better genome partitioning
Luo, R, Sedlazeck, FJ, Darby, CA, Kelly, SM, Schatz, MC (2017) bioRxivdoi: https://doi.org/10.1101/103549 |
 |
|
| P5. |
SplitThreader: Exploration and analysis of rearrangements in cancer genomes
Nattestad, M, Alford, MC, Sedlazeck, FJ, Schatz, MC (2016) bioRxivdoi: https://doi.org/10.1101/087981 |
 |
|
| P4. |
Ribbon: Visualizing complex genome alignments and structural variation
Nattestad, M, Chin, CS, Schatz, MC (2016) bioRxivdoi: http://dx.doi.org/10.1101/082123 |
 |
|
| P3. |
The DOE Systems Biology Knowledgebase (KBase)
Arkin, AP, Steven, RL, Cottingham, RW, Maslov, S, Henry, CS, et al. (2016) bioRxiv doi: https://doi.org/10.1101/096354 |
 |
|
| P2. |
Third-generation sequencing and the future of genomics
Lee, H, Gurtowski, J, Yoo, S, Nattestad, M, Marcus, S, Goodwin, S, McCombie, WR Schatz, MC (2016) bioRxiv doi: http://dx.doi.org/10.1101/048603 |
 |
|
| P1. |
Error correction and assembly complexity of single molecule sequence reads.
Lee, H, Gurtowski, J, Yoo, S, Marcus, S, McCombie, WR, Schatz MC (2014) bioRxiv doi: http://dx.doi.org/10.1101/006395 |
 |
|
|
|
2017
|
| 88. |
16GT: a fast and sensitive variant caller using a 16-genotype probabilistic model
Luo, R, Schatz, MC, Salzberg, SL (2017) GigaScience doi: https://doi.org/10.1093/gigascience/gix045 |
 |
|
| 87. |
Recurrent noncoding regulatory mutations in pancreatic ductal adenocarcinoma
Feigin, ME, Garvin, T, Bailey, P, Waddell, N, Chang, DK, Kelley, DR, Shuai, S, Gallinger, S, McPherson, JD,
Grimmond, SM, Khurana, E, Stein, LD, Biankin, AV, Schatz, MC, Tuveson, DA (2017) Nature Genetics doi:10.1038/ng.3861 |
 |
|
|
| 86. |
GenomeScope: Fast reference-free genome profiling from short reads
Vurture, GW, Sedlazeck, FJ, Nattestad, M, Underwood, CJ, Fang, H, Gurtowski, J, Schatz, MC (2017) Bioinformatics doi: https://doi.org/10.1093/bioinformatics/btx153 |
 |
|
|
| 85. |
Nanopore sequencing meets epigenetics
Schatz, MC (2017) Nature Methods 14, 347–348 (2017) doi:10.1038/nmeth.4240 |
 |
|
|
| 84. |
Bioinformatics of DNA
Heath, LS, Bravo, HC, Caccamo, M, Schatz, MC (2017) Proceedings of the IEEE doi: 10.1109/JPROC.2017.2652958
See entire issue here |
 |
|
|
| 83. |
TGF-β reduces DNA ds-break repair mechanisms to heighten genetic diversity and adaptability of CD44+/CD24- cancer cells
Pal, D, Pertot, A, Shirole, NH, Yao, Z, Anaparthy, N, Garvin, T, Cox, H, Chang, K, Rollins, F, Kendall, J, Edwards, L, Singh, VA,
Stone, GC, Schatz, MC, Hicks, J, Hannon, G, Sordella, R (2017) eLife doi: http://dx.doi.org/10.7554/eLife.21615 |
 |
|
|
2016
|
| 82. |
NanoBLASTer: Fast alignment and characterization of Oxford Nanopore single molecule sequencing reads
Amin, M.R., Skiena, S, Schatz, MC (2016) Proceedings of the Computational Advances in Bio and Medical Sciences (ICCABS) Conference. doi: 10.1109/ICCABS.2016.7802776 |
 |
|
| 81. |
Indel variant analysis of short-read sequencing data with Scalpel
Fang, H, Grabowska, EA, Arora, K, Vacic, V, Zody, MC, Iossifov, I, ORawe, JA, Wu, Y, Jimenez-Barron, LT, Rosenbaum, J, Ronemus, M, Lee, Y, Wang, Z, Lyon, GJ, Wigler, M, Schatz, MC , Narzisi G (2015) Nature Protocols doi:10.1038/nprot.2016.150 |
 |
|
| 80. |
The evolution of inflorescence diversity in the nightshades and heterochrony during meristem maturation
Lemmon, ZH, Park, SJ, Jiang, K, Van Eck, J, Schatz, MC, Lippman ZB (2016) Genome Research doi: 10.1101/gr.207837.116 |
 |
|
| 79. |
Phased Diploid Genome Assembly with Single Molecule Real-Time Sequencing
Chin, CS, Peluso, P, Sedlazeck, FJ, Nattestad, M, Concepcion, GT, Clum, A, Dunn, C, O'Malley, R, Figueroa-Balderas, R, Morales-Cruz, A, Cramer, GR, Delledonne, M, Luo, C, Ecker, JR, Cantu, D, Rank, DR., Schatz, MC (2016) Nature Methods doi:10.1038/nmeth.4035 |
 |
|
| 78. |
Complete telomere-to-telomere de novo assembly of the Plasmodium falciparum genome through long-read (>11 kb), single molecule, real-time sequencing
Vembar, SS, Seetin, M, Lambert, C, Nattestad, M, Schatz, MC, Baybayan, P, Scherf, A, Smith, ML. (2016) DNA Research doi: 10.1093/dnares/dsw022 |
 |
|
| 77. |
Assemblytics: a web analytics tool for the detection of variants from an assembly
Nattestad, M, Schatz, MC (2016) Bioinformatics doi: 10.1093/bioinformatics/btw369 |
 |
|
| 76. |
Insight into the evolution of the Solanaceae from the parental genomes of Petunia hybrida
Bombarely, A et al. (2016) Nature Plants doi:10.1038/nplants.2016.74 |
 |
|
| 75. |
Chromosomal-Level Assembly of the Asian Seabass Genome Using Long Sequence Reads and Multi-layered Scaffolding
Vij, S et al. (2016) PLOS Genetics http://dx.doi.org/10.1371/journal.pgen.1005954 |
 |
|
| 74. |
Genome assembly and geospatial phylogenomics of the bed bug Cimex lectularius
Rosenfeld, JA et al. (2016) Nature Communication doi:10.1038/ncomms10164 |
 |
|
2015
|
| 73. |
The pineapple genome and the evolution of CAM photosynthesis
Ming, R, VanBuren, R, Man Wai, C, Tang, H, Schatz MC, et al. (2015) Nature Genetics doi:10.1038/ng.3435 |
 |
|
| 72. |
Teaser: Individualized benchmarking and optimization of read mapping results for NGS data
Smolka M, Rescheneder P, Schatz MC, Haeseler AV, Sedlazeck FJ. (2015) Genome Biology 16:235 doi:10.1186/s13059-015-0803-1 |
 |
|
| 71. |
Oxford Nanopore sequencing, hybrid error correction, and de novo assembly of a eukaryotic genome
Goodwin, S, Gurtowski, J, Ethe-Sayers, S, Deshpande, P, Schatz MC*, McCombie, WR* (2015) Genome Research doi: 10.1101/gr.191395.115 |
 |
|
| 70. |
Biological data sciences in genome research
Schatz MC (2015) Genome Research 25: 1417-1422. doi: 10.1101/gr.191684.115 |
 |
|
| 69. |
Metassembler: merging and optimizing de novo genome assemblies
Wences, AH Schatz MC (2015) Genome Biology 16:207. doi:10.1186/s13059-015-0764-4 |
 |
|
| 68. |
Genome and transcriptome of the regeneration-competent flatworm, Macrostomum lignano
Wasik, KA, Gurtowski J, Zhou, X, Mendivil Ramos O, Delás MJ, Battistoni, G, El Demerdash, O, Faciatori, I, Vizoso, DV,
Ladurner, P, Schärer, L, McCombie, WR, Hannon, GJ*, Schatz MC* (2015) PNAS doi: 10.1073/pnas.1516718112 |
 |
|
| 67. |
Interactive analysis and assessment of single-cell copy-number variations.
Garvin, T, Aboukhalil, R, Kendall, J, Baslan, T, Atwal, GS, Hicks, J, Wigler, M, Schatz MC (2015) Nature Methods doi:10.1038/nmeth.3578 |
 |
|
| 66. |
Dual functions of Macpiwi1 in transposon silencing and stem cell maintenance in the flatworm Macrostomum lignano
Zhou X, Battistoni G, Demerdash OE, Gurtowski J, Wunderer J, Falciatori I, Ladurner P, Schatz MC, Hannon GJ, Wasik KA (2015) RNA doi: 10.1261/rna.052456.115 |
 |
|
| 65. |
Big Data: Astronomical or Genomical?
Stephens, Z, et al. (2015) PLOS Biology DOI: 10.1371/journal.pbio.1002195 |
 |
|
| 64. |
Molecular genetic diversity and characterization of conjugation genes in the fish parasite Ichthyophthirius multifiliis
MacColl E, et al. (2015) Molecular Phylogenetics and Evolution 86:1-7. doi:10.1016/j.ympev.2015.02.017 |
 |
|
| 63. |
Extending reference assembly models
Church, DM, et al. (2015) Genome Biology 16:13 doi:10.1186/s13059-015-0587-3 |
 |
|
| 62. |
The challenge of small-scale repeats for indel discovery
Narzisi, G, Schatz MC (2015) Frontiers in Bioengineering and Biotechnology 3:8. doi: 10.3389/fbioe.2015.00008 |
 |
|
|
|
2014
|
| 61. |
Whole genome de novo assemblies of three divergent strains of rice (O. sativa) documents novel gene space of aus and indica
Schatz MC, Maron, LG, Stein, JC, Wences, AH, Gurtowski, J, Biggers, E, Lee, H, Kramer, M, Antoniou, E, Ghiban, E, Wright, MH, Chia, JM, Ware, D, McCouch, S, McCombie, WR (2014) Genome Biology 15:506 doi:10.1186/s13059-014-0506-z |
 |
|
| 60. |
SplitMEM: A graphical algorithm for pan-genome analysis with suffix skips
Marcus, S, Lee, H, Schatz MC (2014) Bioinformatics doi: 10.1093/bioinformatics/btu756 |
 |
|
| 59. |
The contribution of de novo coding mutations to autism spectrum disorder
Iossifov, I et al. (2014) Nature doi:10.1038/nature13908 |
 |
|
| 58. |
Reducing INDEL calling errors in whole-genome and exome sequencing data
Fang, H, Narzisi, G, O'Rawe, J, Wu, Y, Rosenbaum, J, Ronemus, M, Iossifov, I, Schatz MC, Lyon, GJ (2014) Genome Medicine 6:89. doi:10.1186/s13073-014-0089-z |
 |
|
| 57. |
Accurate de novo and transmitted indel detection in exome-capture data using microassembly.
Narzisi, G, O'Rawe, JA, Iossifov, I, Fang, H, Lee, YH, Wang, Z, Wu, Y, Lyon, G, Wigler, M, Schatz MC (2014) Nature Methods doi:10.1038/nmeth.3069 |
 |
|
| 56. |
High-coverage sequencing and annotated assemblies of the budgerigar genome
Ganapathy, G et al. (2014) GigaScience 3:11 doi:10.1186/2047-217X-3-11 |
 |
|
| 55. |
Large-scale sequencing and assembly of cereal genomes using Blacklight
Blood, P, Marcus, S, Schatz MC (2014) XSEDE14 Atlanta GA, July 13-18. 2014 [preprint] |
 |
|
| 54. |
On Algorithmic Complexity of Biomolecular Sequence Assembly Problem
Narzisi, G, Mishra, B, Schatz MC (2014) Algorithms for Computational Biology Lecture Notes in Computer Science, Vol. 8542 [preprint] |
 |
|
2013
|
| 53. |
Cultivation and Complete Genome Sequencing of Gloeobacter kilaueensis sp. nov., from a Lava Cave in Kīlauea Caldera, Hawai'i
Saw JHW, Schatz M, Brown MV, Kunkel DD, Foster JS, et al. (2013) PLoS One 8(10): e76376. doi:10.1371/journal.pone.0076376 |
 |
|
| 52. |
Assemblathon 2: evaluating de novo methods of genome assembly in three vertebrate species
Bradnam, KR. et al (2013) GigaScience 2:10 doi:10.1186/2047-217X-2-10 |
 |
|
| 51. |
The advantages of SMRT sequencing
Roberts, RJ, Carneiro, MO, and Schatz, MC (2013) Genome Biology 14:405 doi:10.1186/gb-2013-14-6-405 |
 |
|
| 50. |
The DNA60IFX Contest
Schatz, MC, Taylor, J and Schelhorn, S (2013) Genome Biology 14:124 doi:10.1186/gb-2013-14-6-124 |
 |
|
| 49. |
The DNA Data Deluge
Schatz, MC and Langmead, B (2013) IEEE Spectrum July, 2013 |
 |
|
| 48. |
Genome of the long-living sacred lotus (Nelumbo nucifera Gaertn.)
Ming, R et al. (2013) Genome Biology 14:R41 |
 |
|
| 47. |
Sixty years of genome biology
Doolittle, WF et al. (2013) Genome Biology 14:113 |
 |
|
| 46. |
Aluminum tolerance in maize is associated with higher MATE1 gene copy number
Maron, LG et al. (2013) PNAS doi: 10.1073/pnas.1220766110 |
 |
|
2012
|
| 45. |
Computational thinking in the era of big data biology
Schatz, MC (2012) Genome Biology 13:177 |
 |
|
| 44. |
Answering the demands of digital genomics
Titmus, MA, Gurtowski, J, Schatz, MC (2012) Concurrency and Computation: Practice and Experience DOI: 10.1002/cpe.2925 |
 |
|
| 43. |
De novo identification of 'heterotigs' towards accurate and in-phase assembly
of complex plant genomes
Price J, et al. (2012) Proceedings of BIOCOMP'12 Las Vegas, NV. [preprint] |
 |
|
| 42. |
Genotyping in the Cloud with Crossbow
Gurtowski, J, Schatz, MC, Langmead, B (2012) Current Protocols in Bioinformatics 39:15.3.1-15.3.15 |
 |
|
| 41. |
The rise of a digital immune system
Schatz, MC, Phillippy, AM (2012) GigaScience 1:4 |
 |
|
| 40. |
Hybrid error correction and de novo assembly of single-molecule sequencing reads
Koren, S, Schatz, MC, Walenz, BP, Martin, J, Howard, JT, Ganapathy, G, Wang, Z, Rasko, DA, McCombie, WR, Jarvis, ED, Phillippy, AM (2012) Nature Biotechnology. doi:10.1038/nbt.2280 |
 |
|
| 39. |
Genomic Dark Matter: The reliability of short read mapping illustrated by the Genome Mappability Score
Lee, H, Schatz, MC (2012) Bioinformatics. 10.1093/bioinformatics/bts330 |
 |
|
| 38. |
Current challenges in de novo plant genome sequencing and assembly
Schatz, MC, Witkowski, J, McCombie, WR (2012) Genome Biology. 12:243 |
 |
|
| 37. |
De novo gene disruptions in children on the autism spectrum
Iossifov, I. et al. (2012) Neuron. 74:2 285-299 |
 |
|
2011
|
| 36. |
Rate of meristem maturation determines inflorescence architecture in tomato
Park, SJ, Jiang, K, Schatz, MC, Lippman, ZB (2011) PNAS. doi: 10.1073/pnas.1114963109 |
 |
|
| 35. |
Hawkeye and AMOS: visualizing and assessing the quality of genome assemblies
Schatz, MC, et al. (2011) Briefings in Bioinformatics. doi: 10.1093/bib/bbr074 |
 |
|
| 34. |
GAGE: A critical evaluation of genome assemblies and assembly algorithms
Salzberg, SL, et al. (2011) Genome Research. doi: 10.1101/gr.131383.111 |
 |
|
| 33. |
Complex microbiome underlying secondary and primary metabolism in the tunicate-Prochloron symbiosis.
Donia, MS, et al. (2011) PNAS. doi: 10.1073/pnas.1111712108 |
 |
|
| 32. |
Assemblathon 1: A competitive assessment of de novo short read assembly methods.
Earl, DA, et al. (2011) Genome Research. doi: 10.1101/gr.126599.111 |
 |
|
| 31. |
Two new complete genome sequences offer insight into host and tissue specificity of plant pathogenic Xanthomonas spp.
Bogdanove, AJ, et al. (2011) Journal of Bacteriology. doi:10.1128/JB.05262-11 |
 |
|
| 30. |
Rapid Parallel Genome Indexing with MapReduce.
Menon, RK, Bhat, GP, Schatz, MC (2011) Proceedings of the 2nd International Workshop on MapReduce and its applications. HPDC'11, San Jose, CA. [preprint] |
 |
|
2010
|
| 29. |
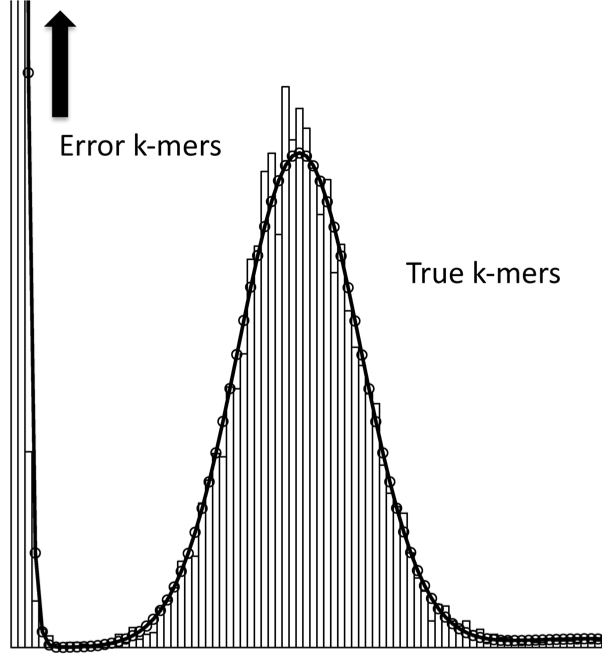
Quake: quality-aware detection and correction of sequencing reads.
Kelley, DR, Schatz, MC, Salzberg, SL (2010) Genome Biology. 11:R116 |
 |
|
| 28. |
Genomic survey of the ectoparasitic mite Varroa destructor, a major pest of the honey bee Apis mellifera.
Cornman, SR, Schatz, MC, et al. (2010) BMC Genomics. 11:602 |
 |
|
| 27. |
Multi-Platform Next-Generation Sequencing of the Domestic Turkey (Meleagris gallopavo): Genome Assembly and Analysis.
Dalloul, RA, et al. (2010) PLoS Biology. 8(9): e1000475. |
 |
|
| 26. |
The missing graphical user interface for genomics.
Schatz, MC. (2010) Genome Biology. 11:128 |
 |
|
| 25. |
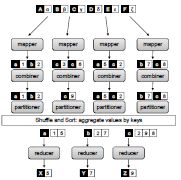
Design patterns for efficient graph algorithms in MapReduce.
Lin, J., Schatz, MC. (2010) Proceedings of the Eighth Workshop on Mining and Learning with Graphs Workshop (MLG-2010) . |
 |
|
| 24. |
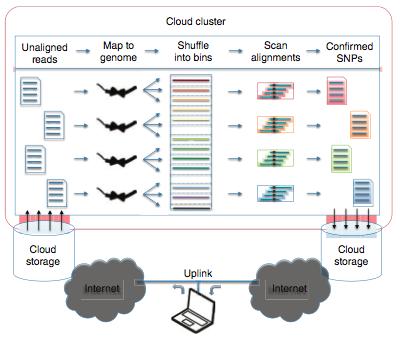
Cloud Computing and the DNA Data Race.
Schatz, MC, Landmead, B, Salzberg, SL. (2010) Nature Biotechnology. 28:691-693 |
 |
|
| 23. |
Integrated microbial survey analysis of prokaryotic communities for the PhyloChip microarray.
Schatz, MC, Phillippy, AM, Gajer, P, DeSantis, TZ, Anderson, GL, Ravel, J. (2010) Applied and Environmental Microbiology. |
 |
|
| 22. |
Assembly of large genomes using second-generation sequencing.
Schatz, MC, Delcher, AL, Salzberg, SL. (2010) Genome Research, 20: 1165-1173. |
 |
|
| 21. |
Assembly complexity of prokaryotic genomes using short reads.
Kingsford, C, Schatz, MC, Pop, M (2010) BMC Bioinformatics 11:21. |
 |
|
2009
|
| 20. |
Searching for SNPs with cloud computing.
Langmead, B, Schatz, MC, Lin, J, Pop, M, Salzberg, SL (2009) Genome Biology 10:R134. |
 |
|
| 19. |
Optimizing data intensive GPGPU computations for DNA sequence alignment.
Trapnell, C, Schatz, MC (2009) Parallel Computing 35(8-9):429-440. |
 |
|
| 18. |
Genomic Analyses of the Microsporidian Nosema ceranae, an Emergent Pathogen of Honey Bees.
Cornman, RS, Chen, YP, Schatz, MC, et al. (2009) PLoS Pathogens 5(6):e1000466. |
 |
|
| 17. |
A whole-genome assembly of the domestic cow, Bos taurus.
Zimin, AV, Delcher, AL, Florea, L, Kelley, DR, Schatz, MC, et al. (2009) Genome Biology 10:R42. |
 |
|
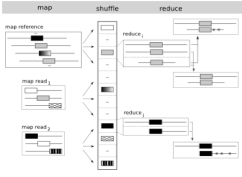
| 16. |
CloudBurst: Highly Sensitive Read Mapping with MapReduce.
Schatz, MC (2009) Bioinformatics 25:1363-1369. |
 |
|
| 15. |
Comparative genomics of mutualistic viruses of Glyptapanteles parasitic wasps.
Desjardins, CA, et al. (2009) Genome Biology 9:R183. |
 |
|
2008
|
| 14. |
Characterization of Insertion Sites in Rainbow Papaya, the First Commercialized Transgenic Fruit Crop.
Suzuki, JY, et al. (2008) Tropical Plant Biology 1:293-309. |
 |
|
| 13. |
Revealing Biological Modules via Graph Summarization.
Navlakha, S, Schatz, M, Kingsford, C. (2008) Journal of Computational Biology 16(2): 253-264. |
 |
|
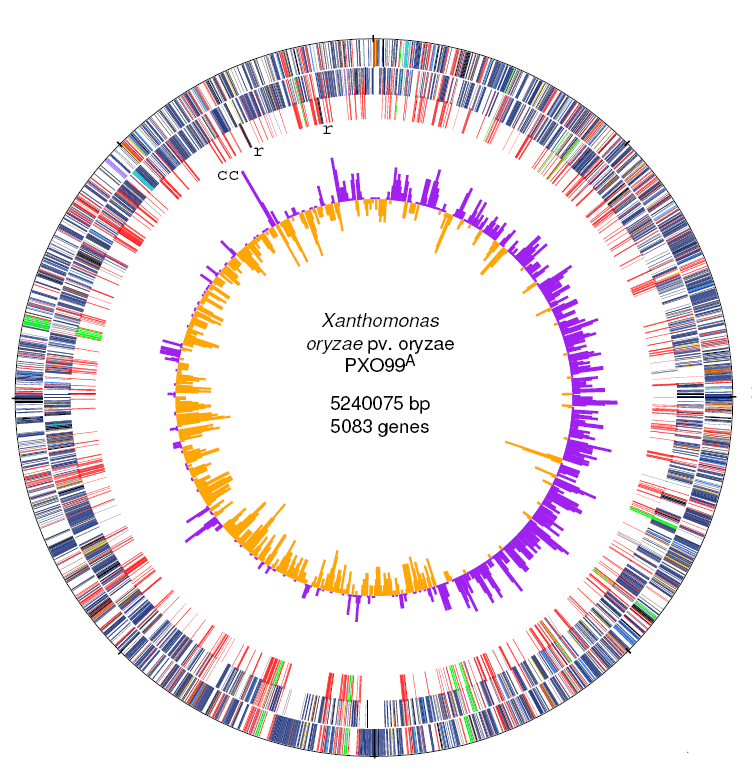
| 12. |
Genome sequence and rapid evolution of the rice pathogen Xanthomonas oryzae pv. oryzae PXO99A.
Salzberg, SL, Sommer DS, Schatz, MC, et al. (2008) BMC Genomics 9:204. |
 |
|
| 11. |
The draft genome of the transgenic tropical fruit tree papaya (Carica papaya Linnaeus).
Ming, R, et al. (2008) Nature 452, 991-996. |
 |
|
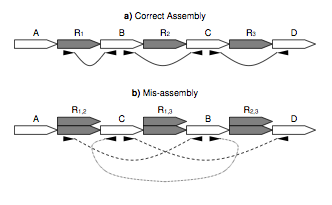
| 10. |
Genome Assembly forensics: finding the elusive mis-assembly.
Phillippy, AM, Schatz, MC, Pop, M. (2008) Genome Biology 9:R55. |
 |
|
2007
|
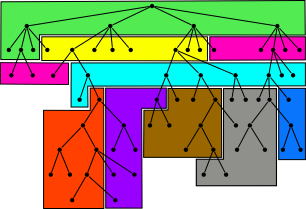
| 9. |
High-throughput sequence alignment using Graphics Processing Units.
Schatz, MC, Trapnell, C, Delcher, AL, Varshney, A. (2007) BMC Bioinformatics 8:474. |
 |
|
| 8. |
Evolution of genes and genomes on the Drosophila phylogeny.
Drosophila 12 Genomes Consortium. (2007) Nature Nov 8;450(7167):203-18. |
 |
|
| 7. |
Draft Genome of the Filarial Nematode Parasite Brugia malayi.
Ghedin, E, et al. (2007) Science 317(5845):1756-1760. |
 |
|
| 6. |
Structure and evolution of a proviral locus of Glyptapanteles indiensis bracovirus.
Desjardins, CA, et al. (2007) BMC Microbiology 7:61 |
 |
|
| 5. |
Genome Sequence of Aedes aegypti, a Major Arbovirus Vector.
Nene, V et al. (2007) Science 316(5832), 1718-1723. |
 |
|
| 4. |
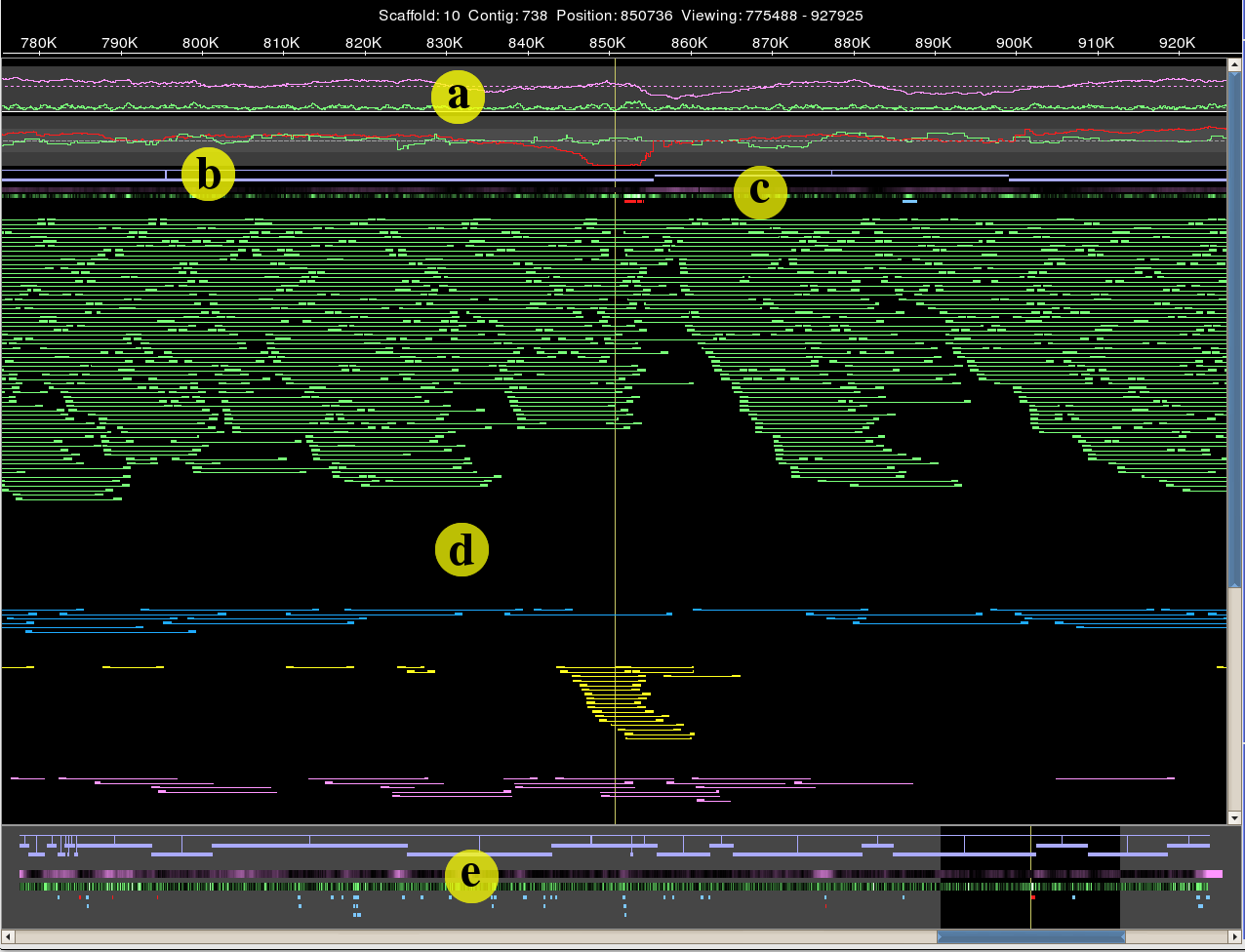
Hawkeye: a visual analytics tool for genome assemblies.
Schatz, MC, Phillippy, AM, Shneiderman, B, Salzberg, SL. (2007) Genome Biology 8:R34. |
 |
|
| 3. |
Draft Genome Sequence of the Sexually Transmitted Pathogen Trichomonas vaginalis.
Carlton, JM, Hirt, RP, Silva, JC, Delcher, AL, Schatz, M, et al. (2007) Science 315 (5809), 207-212. |
 |
|
2006 & earlier
|
| 2. |
Major structural differences and novel potential virulence mechanisms from the genomes of multiple campylobacter species.
Fouts, DE et al. (2005) PLoS Biology 3 (1):e15. |
 |
|
| 1. |
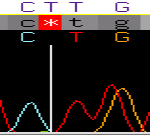
Automated correction of genome sequence errors.
Gajer, P, Schatz, M, Salzberg, SL. (2004) Nucleic Acids Research 32 (2):562-569. |
 |
|
See more in Google Scholar.
|
|
